import torch
from torch.utils.data import DataLoader
import deepinv as dinv
from utils import Trainer, ArtifactRemovalCRNN, CRNN, DeepinvSliceDataset, CineNetDataTransformFully unsupervised dynamic MRI reconstruction via geometrotemporal equivariance
Aim: reconstruct dynamic MRI videos from accelerated undersampled measurements \(\mathbf{y}=\mathbf{Ax}\) where \(\mathbf{A}\) is an undersampled Fourier operator.
Applications: real-time cardiac imaging, free-breathing motion, vocal tract speech…
Goals:
- Capture true motion: aperiodicities, irregularities: real-time MRI
- Capture higher spatiotemporal resolution with fewer measurements (leading to faster, cheaper, portable MRI)
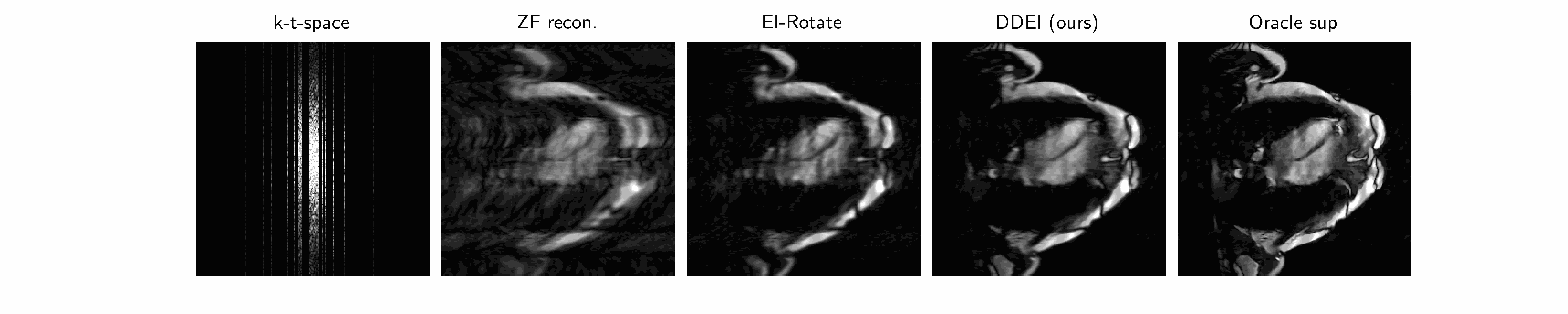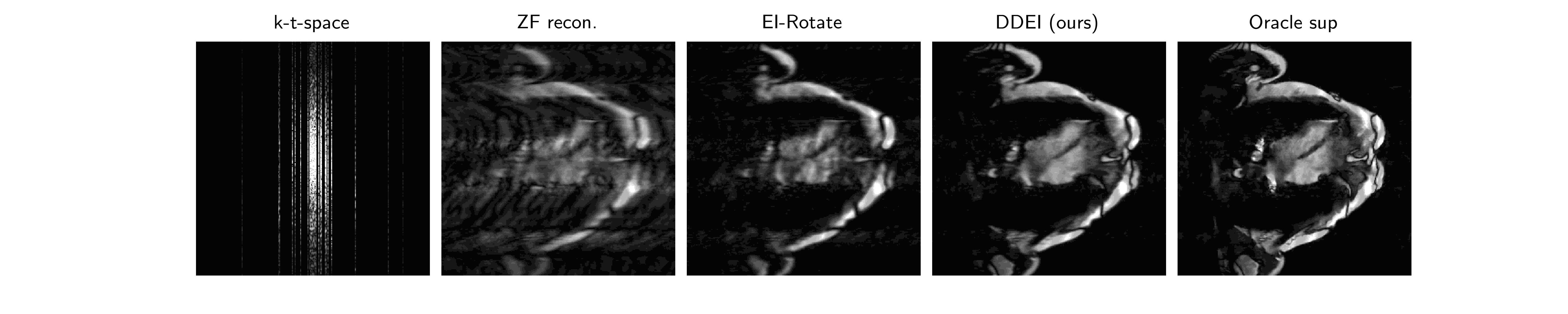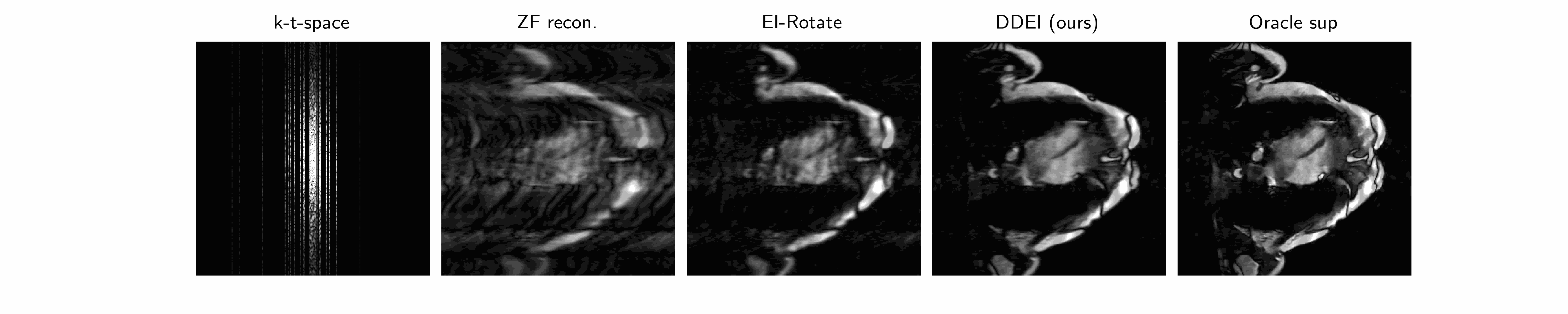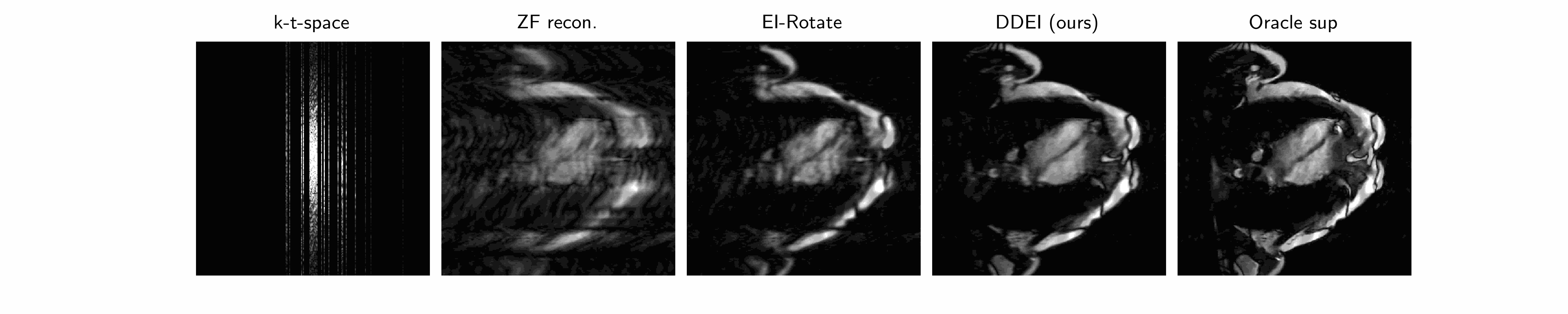
Why is it hard? ground truth is impossible to truly obtain! There is no such thing as true fully-sampled dynamic MRI data at the same frame rate as the measurement data. Hence all supervised methods (CineNet, transformers…) are fundamentally flawed - an implicit data crime. The best pseudo-ground-truth, e.g. gating/cine imaging, must assume periodicity and all methods that use this cannot capture true motion and its irregularities - which is often of interest in medical imaging. Therefore we need unsupervised methods.
Our method we posit that the unknown set \(\mathcal{X}\) of MRI videos is \(G\)-invariant: \(\forall x\in\mathcal{X},g\cdot x\in\mathcal{X}\forall g\in G\). We propose a spatiotemporal group combining temporal invariance and diffeomorphic invariance: see our paper for more details. We use the Equivariant Imaging framework to constrain this with a ground-truth-free loss, which we call Dynamic Diffeomorphic Equivariant Imaging (DDEI).
Results: see below.
You can easily implement our method using the deepinv library. See train.py for a full training demo including training and evaluating competitors. Contents:
Define dynamic MRI physics:
Define accelerated dynamic MRI. We set the (4x, 8x, 16x) undersampling mask on-the-fly as it varies per subject.
physics = dinv.physics.DynamicMRI(img_size=(1, 2, 12, 512, 256), device="cpu")Define the DDEI loss function:
See train.py for full demo of how to train with competitors’ losses using deepinv.
transform = dinv.transform.ShiftTime() | (dinv.transform.CPABDiffeomorphism() | dinv.transform.Rotate())
loss = [dinv.loss.MCLoss(), dinv.loss.EILoss(transform=transform)]Define the data:
We use a real-world cardiac 2D+t dataset from the 2023 CMRxRecon challenge. This dataset has fully-sampled cine measurements which we use as GT for evaluation, but we emphasise our method does not require gated GT for training. We then simulate 2D+t random Cartesian undersampling masks using deepinv, see this notebook for more details. For more details on how to download the full dataset see README.
dataset = DeepinvSliceDataset(
root="data/CMRxRecon",
transform=CineNetDataTransform(time_window=12, apply_mask=True, normalize=True),
set_name="TrainingSet",
acc_folders=["FullSample"],
mask_folder="TimeVaryingGaussianMask08",
dataset_cache_file="cache.pkl"
)
loader = DataLoader(dataset=dataset)100%|██████████| 1/1 [00:00<00:00, 4.81it/s]Saving dataset cache fileDefine neural network:
For \(f_\theta\) we use a very small CRNN, a lightweight unrolled network with 2 unrolled iterations and 1154 parameters. Our framework is NN-agnostic and any state-of-the-art NN can be used as the backbone.
model = ArtifactRemovalCRNN(CRNN(num_cascades=2)).to("cpu")Train the network!
We train the network using a modified deepinv.Trainer. For full training demo, see train.py.
trainer = Trainer(
model = model,
physics = physics,
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3),
train_dataloader = loader,
losses = loss,
metrics = dinv.metric.PSNR(complex_abs=True, max_pixel=None)
)
trainer.train()Test a pretrained model
We load the model used for the paper trained with 50 epochs on our full train set, provided in models/. Test on a sample image included locally and compare to the initial zero-filled reconstruction.
model.load_state_dict(torch.load("models/ddei_pretrained_50.pth.tar", map_location="cpu")["state_dict"])<All keys matched successfully>x, y, mask = next(iter(loader))
physics.update_parameters(mask=mask)
x_hat = model(y, physics)
x_zf = physics.A_adjoint(y)dinv.utils.plot_videos([x, y.clip(-2, 2), x_zf, x_hat], titles=["GT", "k-t-space", "Zero-filled", "DDEI"], display=True)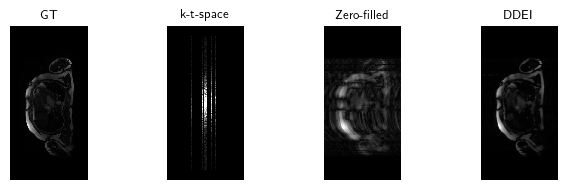
Full results
Test set example cardiac long axis views (above 2 rows) and short axis slice (below) reconstruction results: